Data Version Control 101 with Oxen
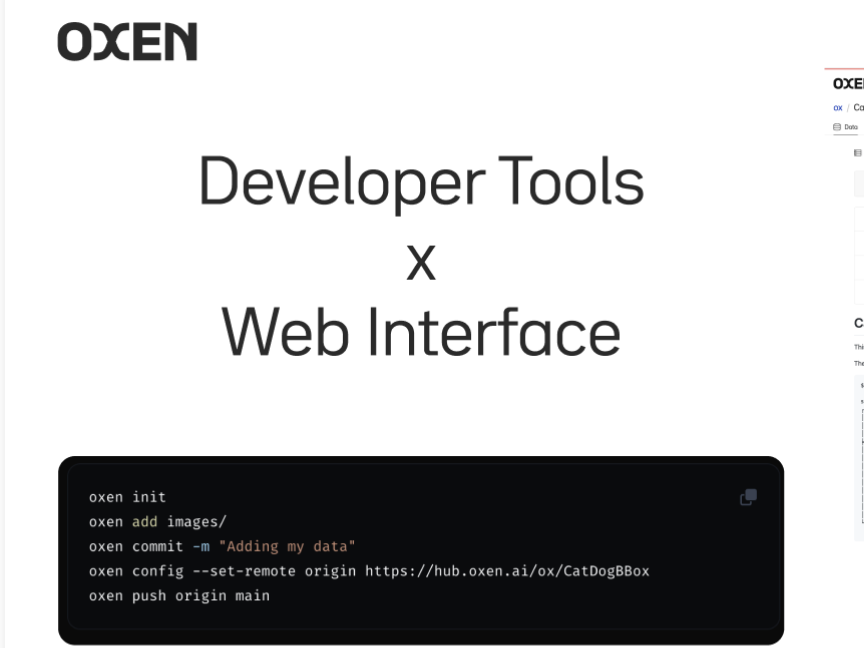
This intro tutorial from Oxen.ai shows how Oxen can make versioning your data as easy as versioning your code.
Oxen is built to track and store changes for everything from a single CSV to data repositories with millions of unstructured images, videos, audio or text files.
The tutorial will go through what data version control is, why it is important, and how Oxen helps data scientists and engineers gain visibility and confidence when sharing data with the rest of their team.
Here's a video version of the same tutorial if following along on YouTube is more your speed.
Why Version Data?
No matter what you job title, we’ve all had the experience of somebody handing you a file of data, and in order to keep track of what version it is, they came up with a clever name. Maybe it was some combination of a name and a date. Maybe it was the second version of that same file, so they added _v2 . Either way, only they truly knew the mystique behind the filename choice.
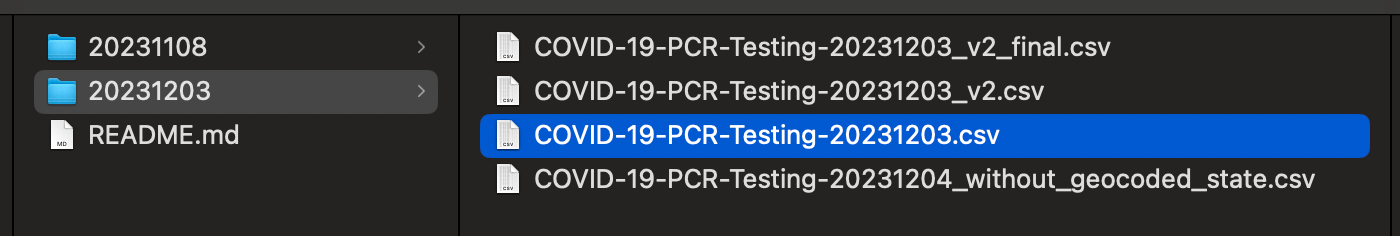
Not only does file naming get out of control, but data changes minute by minute, day by day.
Can you spot the differences between these two datasets?
Version 1
state,name,fips,fema_region,outcome,date,new_reported,total_reported
AL,Alabama,1,Region 4,Negative,2020/03/01,96,96
AL,Alabama,1,Region 4,Positive,2020/03/01,16,16Version 2
state,state_name,state_fips,region,outcome,date,results_reported,results_reported,geocoded_state
AL,Alabama,01,Region 4,Negative,2020/11/05,62,96,
AL,Alabama,01,Region 4,Positive,2020/11/05,14,16,Column names are changed, there is a different distribution of data, some data points might have been filtered or removed. It can be hard to debug and see from just the raw data itself.
All these changes could break downstream analysis or applications that depend it. There is nothing worse as an engineer than having a change in data break your application when model code hasn't changed.
Real World Example
As a concrete example, we will be working with a COVID-19 Diagnostic Testing dataset that we alluded to above. You can imagine this dataset being used to help analyze and predict the spread of COVID-19.
Let's start with version one of the file.
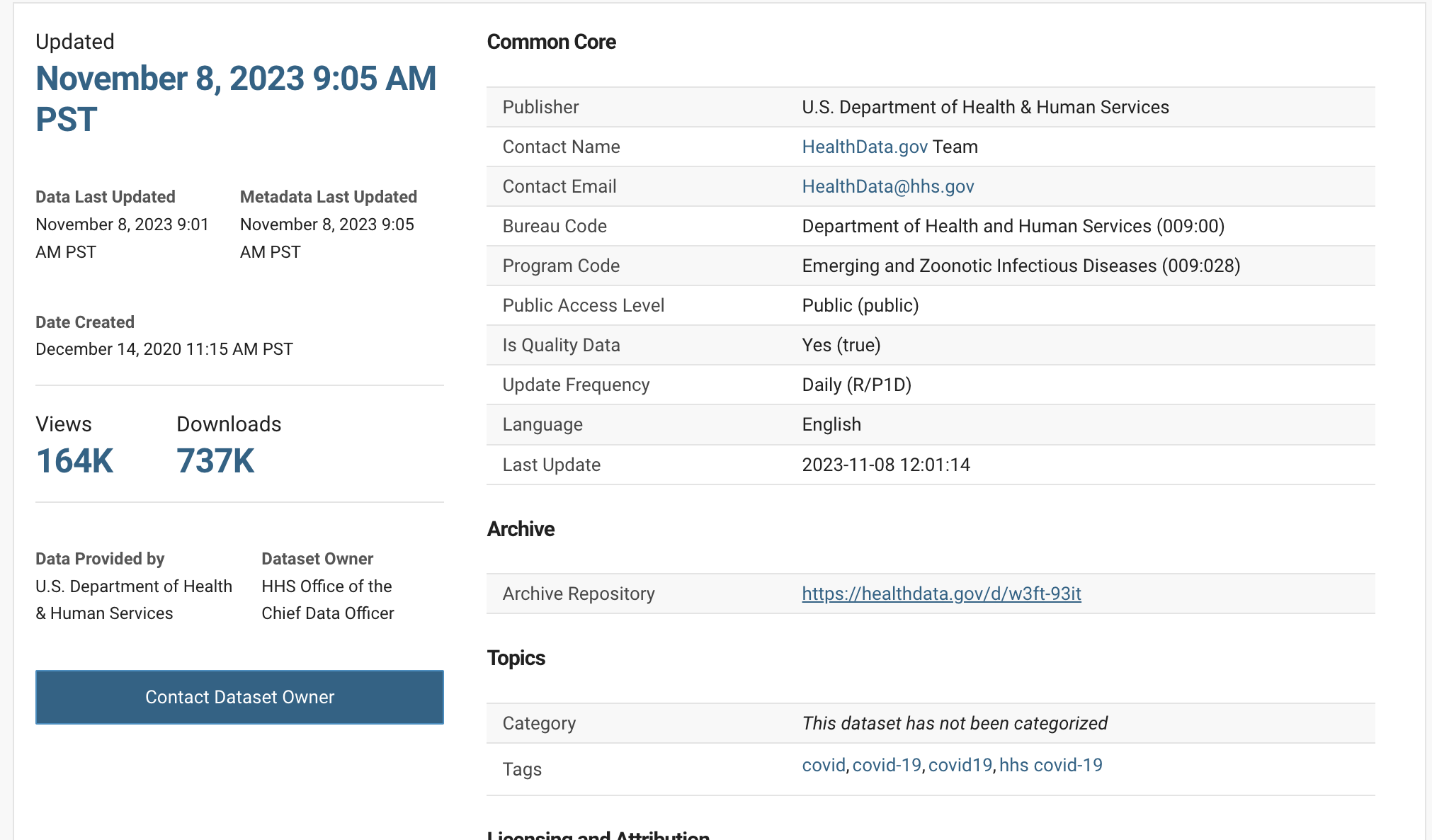
This data was downloaded from HealthData.gov and you can grab your own example from their website.
You can see the initial data was created December 14th, 2020 and it was last updated November 8th, 2023. That’s 3 years of untracked, un-auditable changes that have been lost to the sands of time.
Bad Data Versioning
The lazy way to version your data is some combination of the filename, plus a date, and some other identifying information, so that you can distinguish it from another export of the same data.
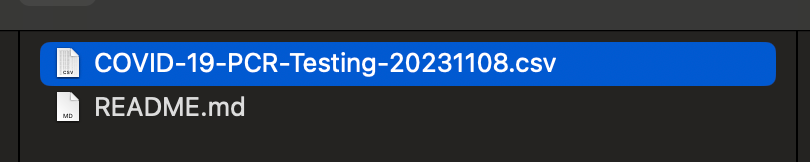
In this case, the person who exported the file decided to use the date of 20231108 as the version in the filename COVID-19-PCR-Testing-20231108.csv .
I went back the next day, and found a new version that was 20231109.
Leave aside sharing data for now. When you name files like this, and come back a day later, let alone a month later, you end up scratching your head as to what the difference is between 20231201 and 20231108 and why there was a _v2 at the end of this random other file.
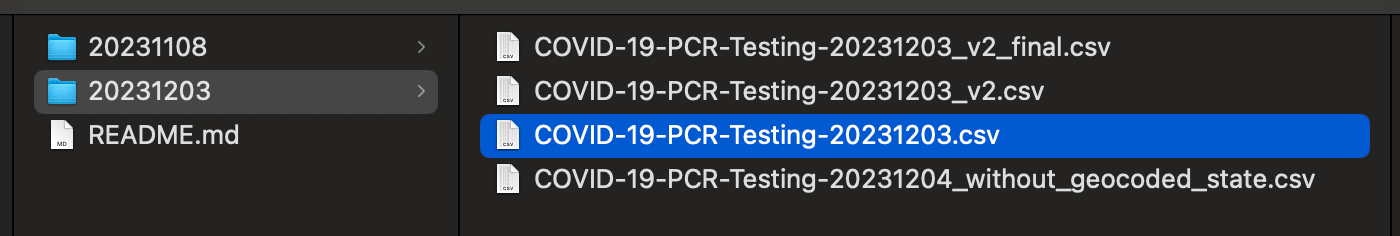
You may argue that the difference is clear in this case....it is the newer set of results with more data! We will see why this assumption is a bit too simplistic later.
Data is not a solo job
Not only are you scratching your head as to what 20231203_v2 vs 20231203_v2_final mean a week later after you had done your data export and cleaning, try to communicate this with the rest of your team.
One teams effort might be to extract and transform the data, while another team’s job is to analyze it, build models, answer questions etc. If the person who exported the file cannot keep the context in their head of which file or directory means what, good luck communicating changes to an external team.
There must be a better way.
Enter Oxen.ai
Oxen is comprised of command line tooling and python libraries for developers, as well as a web interface for anyone who wants access to the data.
The Oxen developer tools are open source and can be found for any developer to get up and running. They can be found on GitHub.
The web interface is a tool that anyone from an engineer, project manager, or data scientist can use without knowing how to code.
Within the developer tools lies a command line interface and python library modeled after git which makes oxen easy to learn for software engineers. The difference is oxen is optimized for datasets instead of code.
How is Oxen Version Control Different?
Oxen is designed from the ground up to handle a variety of workloads. It can any type of data from large CSVS to directories cluttered with images, audio files, or video.
Git and other version control systems were mainly built to handle text files of code, comparing changes line by line.
Sure, there are tools like git-lfs to handle larger files, but they are not a great developer experience for larger datasets and are better for one off files that do not change very often.
Oxen In Your Workflow
We will start with a simple example of a single CSV to wrap our heads around the workflow.
The following assumes you are a developer who is comfortable with the command line. All the examples can also be translated to python if you are more comfortable working in a programming language.
You can find full documentation on how to install oxen in the developer docs. If you are on mac you can simply brew install the command line tools.
brew tap Oxen-AI/oxen
brew install oxenExample Dataset
Let’s say you are trying to analyze the outbreak of COVID-19. Your team is tasked with collecting data and building predictive models to help stop future spreads.
You set up a data pipeline that exports from HealthData.gov databases each night, and pass it over to the data science team to find any patterns and build machine learning models.
Since your team is smart, they have setup Oxen in the loop to help make sure none of the changes are lost, and the whole team has visibility into what is happening.
You can find the starter data in this poorly organized Oxen repository, that we will cleanup.
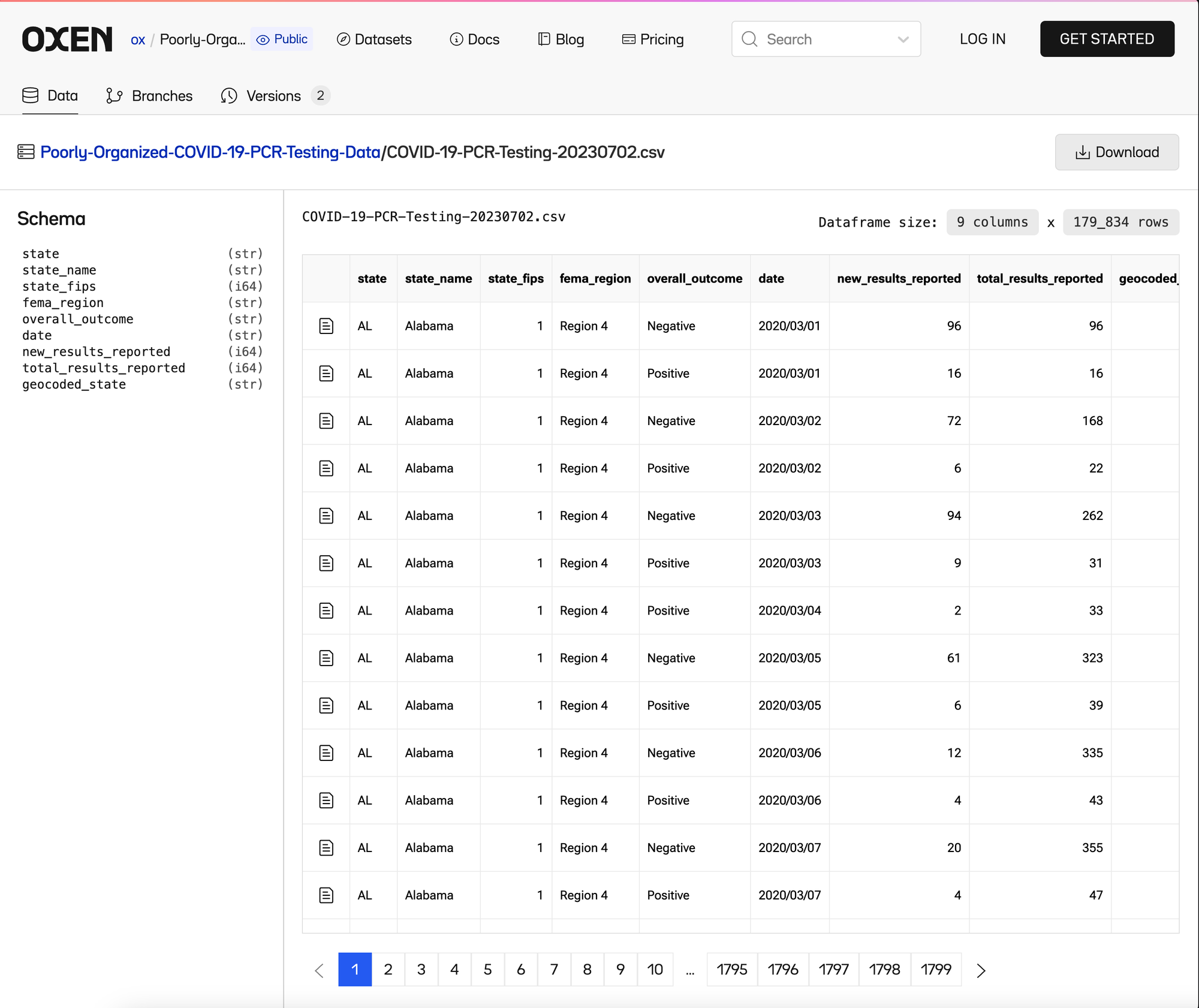
Let’s download the data, and rename the first csv from COVID-19-PCR-Testing-20230702.csv → COVID-19-PCR-Testing.csv
cp COVID-19-PCR-Testing-20230702.csv COVID-19-PCR-Testing.csvNow we can initialize a local Oxen repository and add our data to it.
oxen init
oxen add COVID-19-PCR-Testing.csvJust like git oxen has the concept of commit messages to help keep track of changes.
oxen commit -m "adding the initial data from 2023/07/02"We have now separated the file name from the description from the changes. This allows us to have more verbose messaging and it allows us to track who made the changes and when they were made. Finally we know that COVID-19-PCR-Testing.csv will always be the file we are working with, and no longer have to worry about silly filename conventions.
Share The Data
In order to share this file with our team, or the world, we can use the Oxen Web Interface to create a repository, and push our data there.
If you have not yet, navigate to https://oxen.ai and create an account there. Once your account is made, you can create an empty repository of data.
Next we follow the instructions to configure our API Key, and push the data.
The commands are below to make it easy to copy and paste and replace [YOUR API KEY] with your own found at the top of the repository.
oxen config --auth hub.oxen.ai [YOUR API KEY]
oxen config --set-remote origin https://hub.oxen.ai/ox/COVID-19-PCR-Testing
oxen push origin mainYou will see a little progress bar of our trusty ox pushing your data to the cloud.

Now the data has been pushed to the web hub and anyone with access can view, download, and modify it.
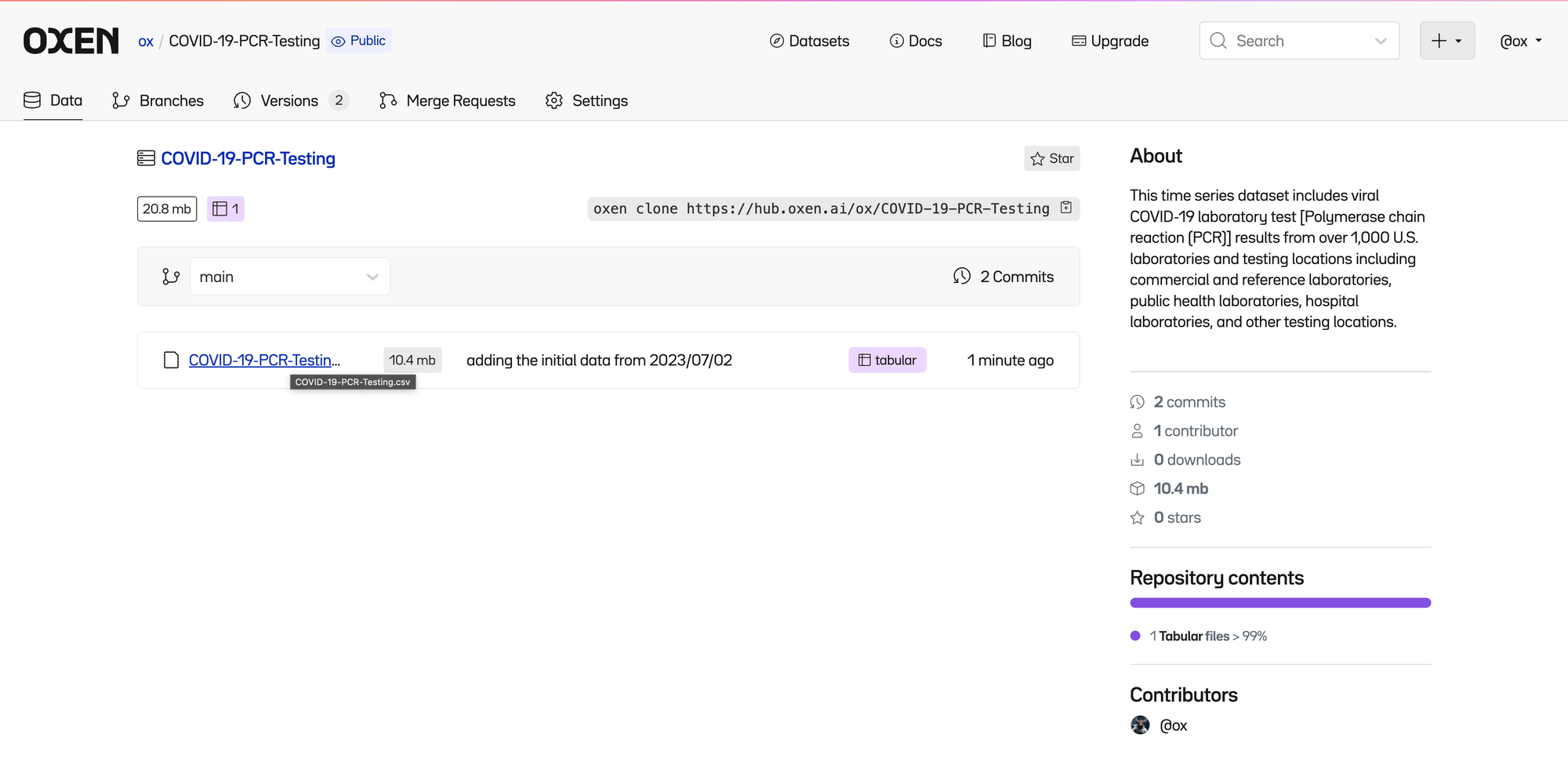
Track Changes To Data
We quickly realize that we do not have the latest data and that there is a new export with data up until 2023/11/08.
One of great things about version control is we can fearlessly overwrite our COVID-19-PCR-Testing.csv with the new data.
Download the newer data from our poorly organized repository and overwrite your file.
cp COVID-19-PCR-Testing-20231108.csv COVID-19-PCR-Testing.csv
oxen statusRunning oxen status we can see Oxen automatically detects that the file was modified.

If we ask Oxen to find the differences between the files, it finds that 92,630 rows were added and 57,215 rows were removed!
oxen diff COVID-19-PCR-Testing.csv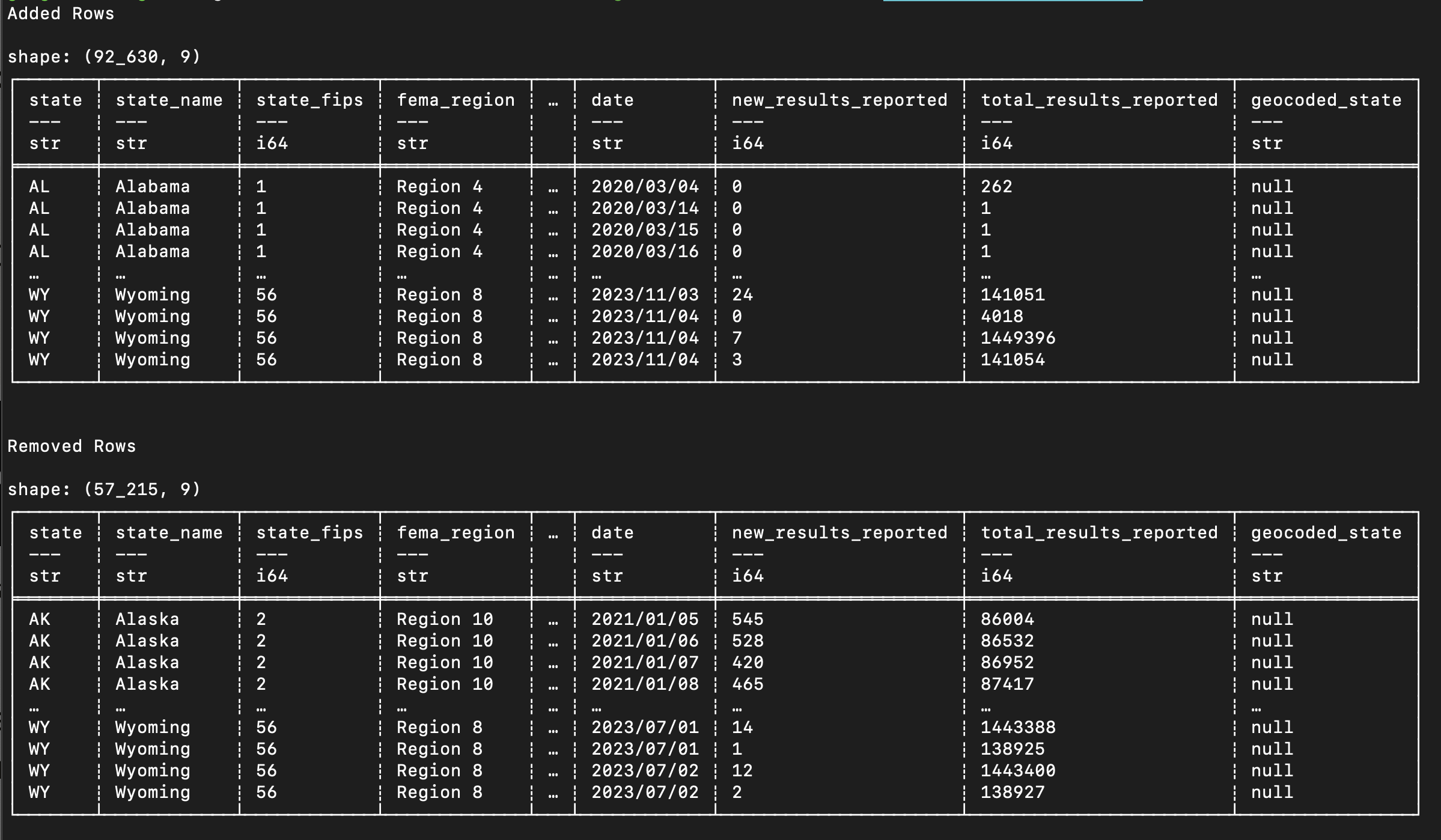
This is a huge change in the data.
Going line by line through these changes might give us some insight, but is clearly not the most efficient way to figure out what changed.
Let’s add and commit these changes, then push them to the web hub to find out more.
oxen add COVID-19-PCR-Testing.csv
oxen commit -m "adding new data from 2023/11/08"
oxen push origin mainAnalyzing Commit History
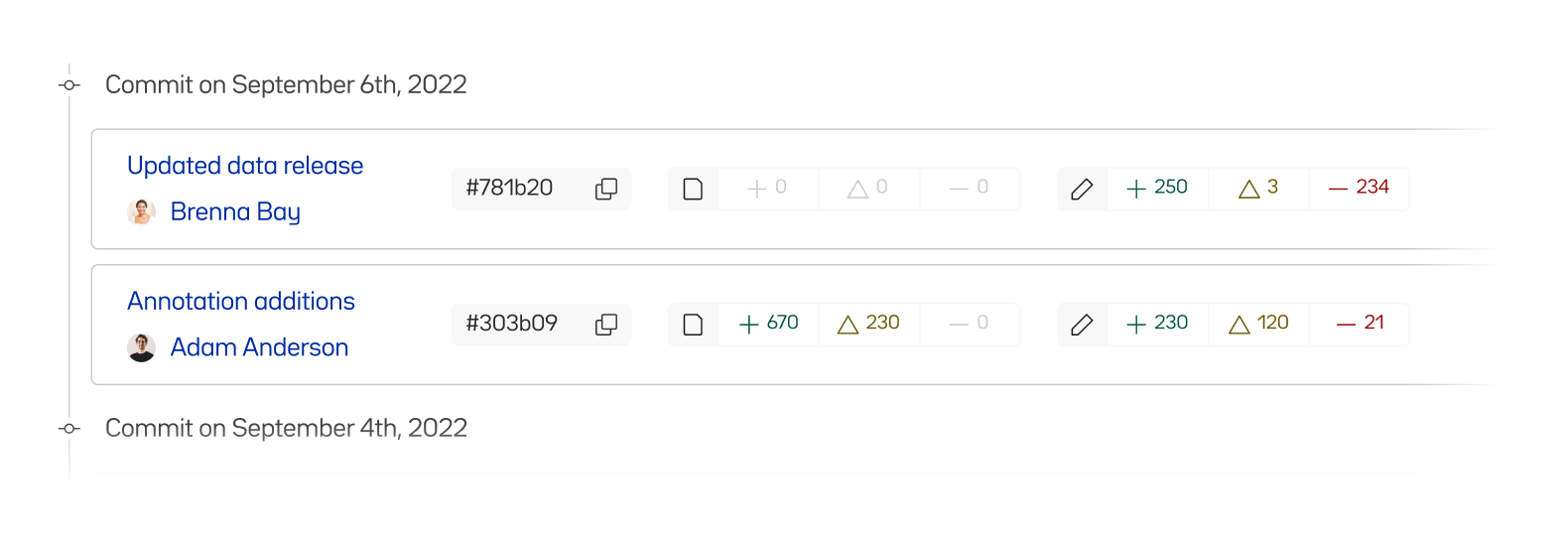
Each time a change is made, it is “committed” to the history. If you click the “3 Commits” link in the upper right of the file list, you can see the list of committed changes as well as who made them.
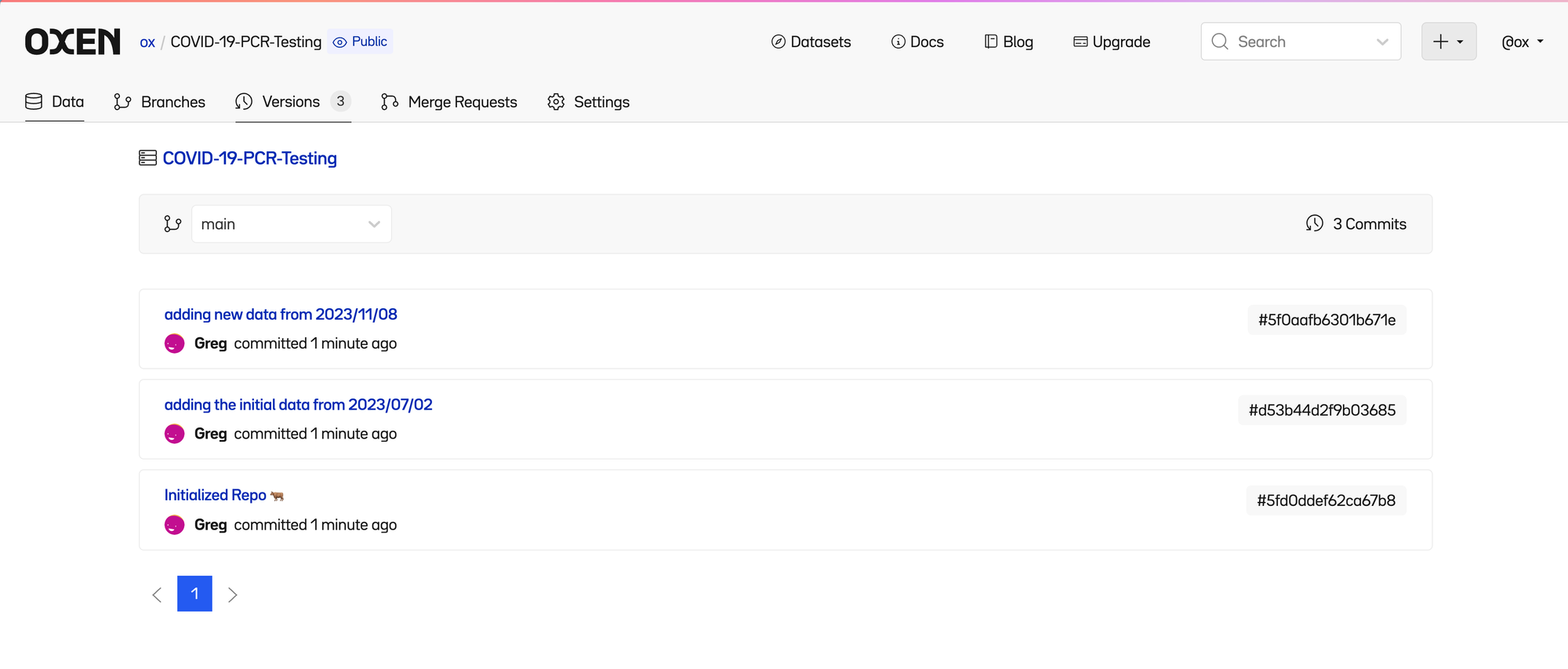
Clicking on the most recent commit lets us see a high level summary of files that were changed.
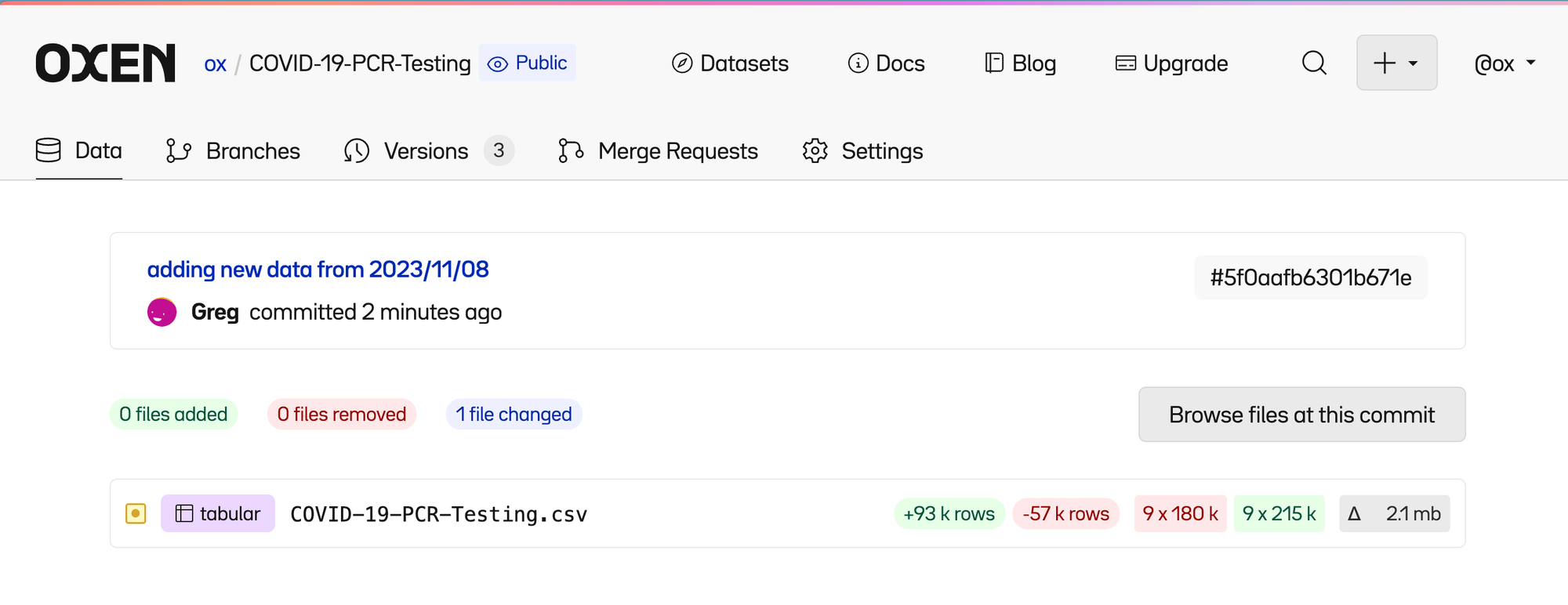
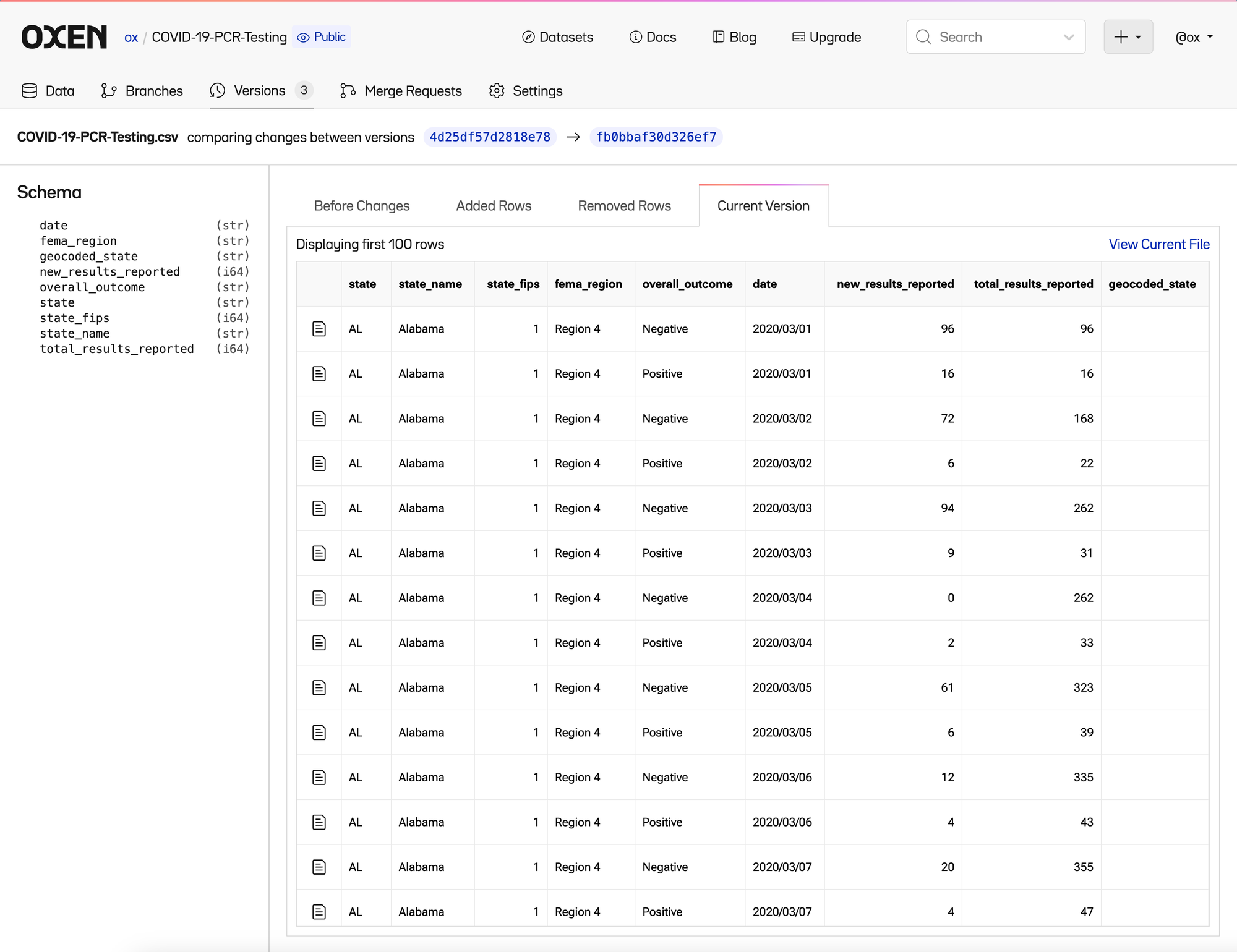
Click into the file and you can see 4 tabs
- Current Version
- Removed Rows
- Added Rows
- Before Changes
All which you can dive into further to figure out what changed between the two versions of the files.
Working in Parallel
Another benefit of versioning the data is being able to work on the same dataset in parallel.
Whether it is a single engineer, or a team of data scientists, everyone has a different idea of how we can process the data and build the best predictive model.
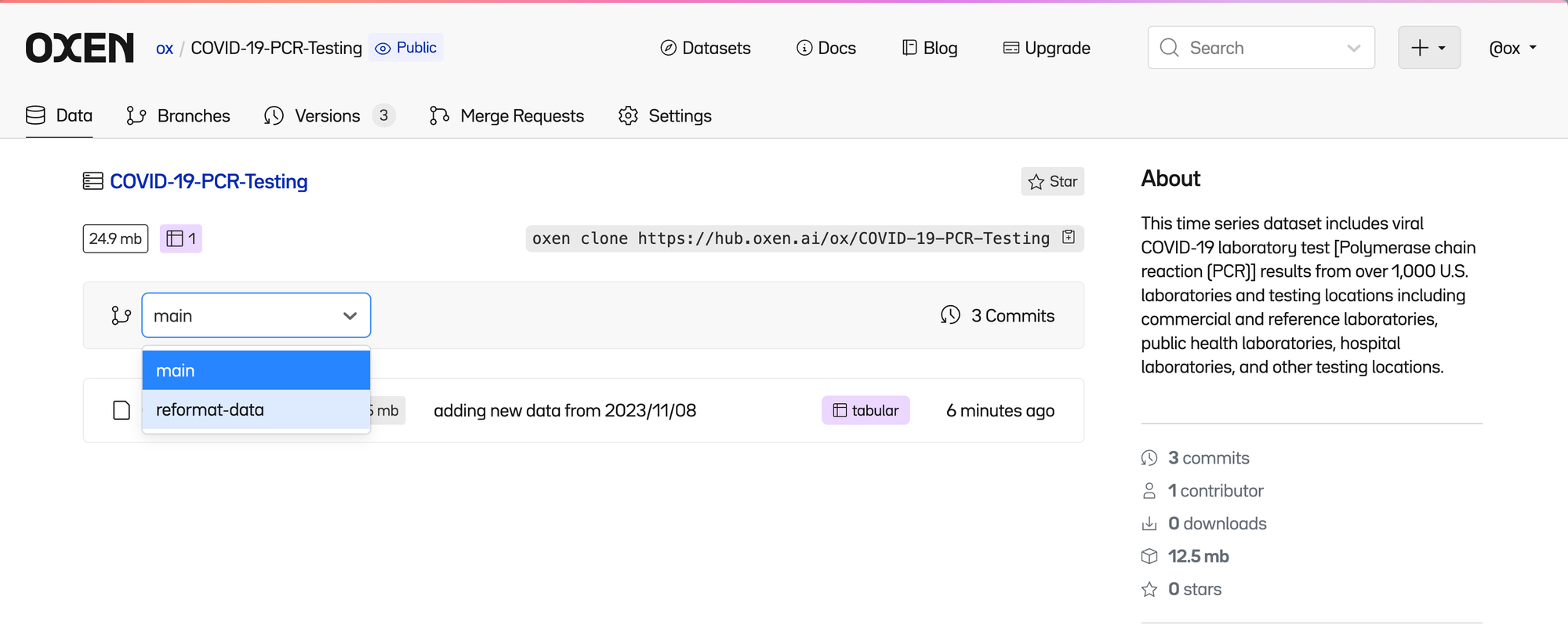
Similar to other version control systems, Oxen has the concept of branches where people can make changes to data without stepping on each other’s toes.
Branches can made in the command line and pushed up to the web hub.
oxen checkout -b reformat-data
oxen push origin reformat-dataI can have my working version of COVID-19-PCR-Testing.csv in a branch called reformat-data you can still have access to the original in the main branch (which is created by default).
You can seamlessly swap between branches in the web interface.
Column Level Changes
Each CSV has an associated "schema" which is the list of column names and data types.
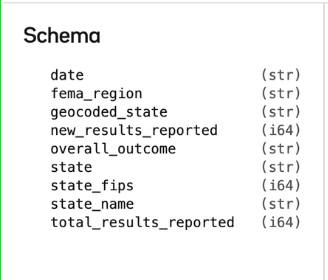
Say that I had a clever new naming convention for our columns that was easier for me to understand. This may break downstream systems, but I think it is going to make things clearer going forward.
Grab an example of these changes here with the poorly named file COVID-19-PCR-Testing-20231108_v2.csv
# overwrite the data with the new formatted data
cp COVID-19-PCR-Testing-20231108_v2.csv COVID-19-PCR-Testing.csv
# stage the changes with oxen
oxen add COVID-19-PCR-Testing.csv
# commit the changes to oxen
oxen commit -m "adding reformatted data from 2023/11/08"
# push the changes to the remote reformat-data branch
oxen push origin reformat-dataAfter pushing the changes to the hub, you now have a parallel history of the data and changes.
Oxen makes it easy to see these changes as they happen, so that we can decide as a team if it is the right direction to pursue. You can see the new column names in the “Current Version”
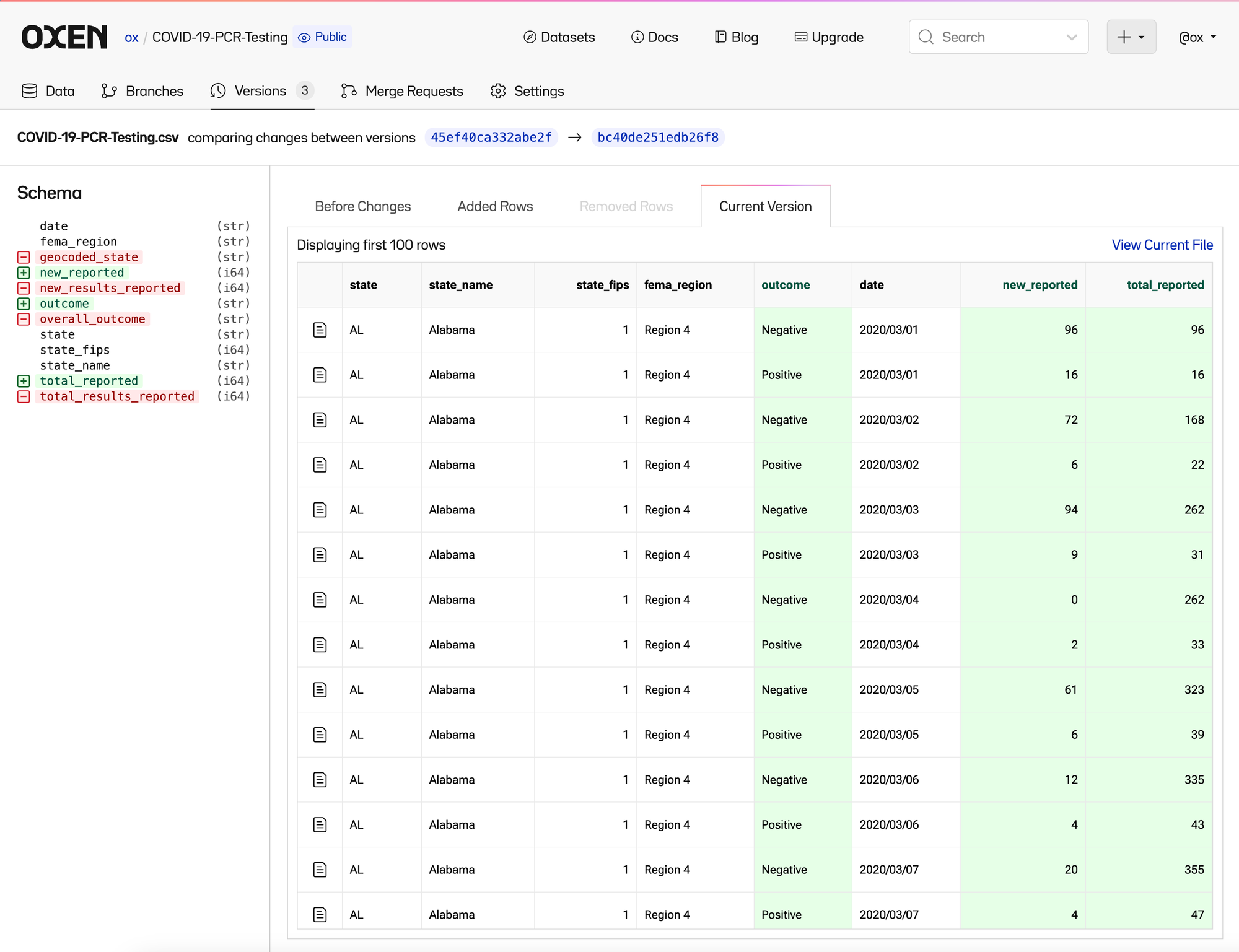
As well as the removed column names in the previous version.
The changes we have made so far are not that complicated, but this is a 101 tutorial.
Even with these simple changes it quickly becomes evident why having version control in the loop while working with a team helps them fearlessly add, modify, transform data all while keeping the entire history of changes in an organized manner.
Download the version you need
Now that you understand the flow for uploading and iterating on different versions, you may be asking how you integrate this into the rest of your workflow.
You can download individual files and directories, given a specific branch or version easily with the user interface by navigating to the file and clicking the "Download" button.
You can also programmatically use the command line interface, python library, or HTTP to download the files.
Command Line
oxen clone http://hub.oxen.ai/ox/CatDogBBox --shallow
cd CatDogBBox
oxen remote download annotations/test.csvPython Library
from oxen import RemoteRepo
repo = RemoteRepo("http://hub.oxen.ai/ox/CatDogBBox")
repo.download("annotations/test.csv")Curl/HTTP
curl -X GET -H "Authorization: Bearer $TOKEN" https://hub.oxen.ai/api/repos/ox/CatDogBBox/file/main/annotations/test.csv -o ~/Downloads/test.csvView the full documentation here.

Now any downstream engineer, data scientist, or application can fetch the version of the file that they know works, and can understand how and why data has changed since the moment they wrote their initial code.
Future you and your future team members will thank you.
Next Steps
Now that you understand what data version control is, and hopefully see the value of it, dive into the many datasets already available on the Oxen Hub.
Whether you are solving a traditional data science problem with time series data or building and evaluating advanced computer vision or large language models, Oxen allows teams to experiment fearlessly and communicate changes effectively.
Create a repository, upload your data, and share with your team a better way!
We'd love to see what you're building with these tools! Reach out to hello@oxen.ai, follow us on Twitter @oxendrove, give us a ⭐ on GitHub, dive deeper into the documentation, or Sign up for Oxen today.


